Muscle Research
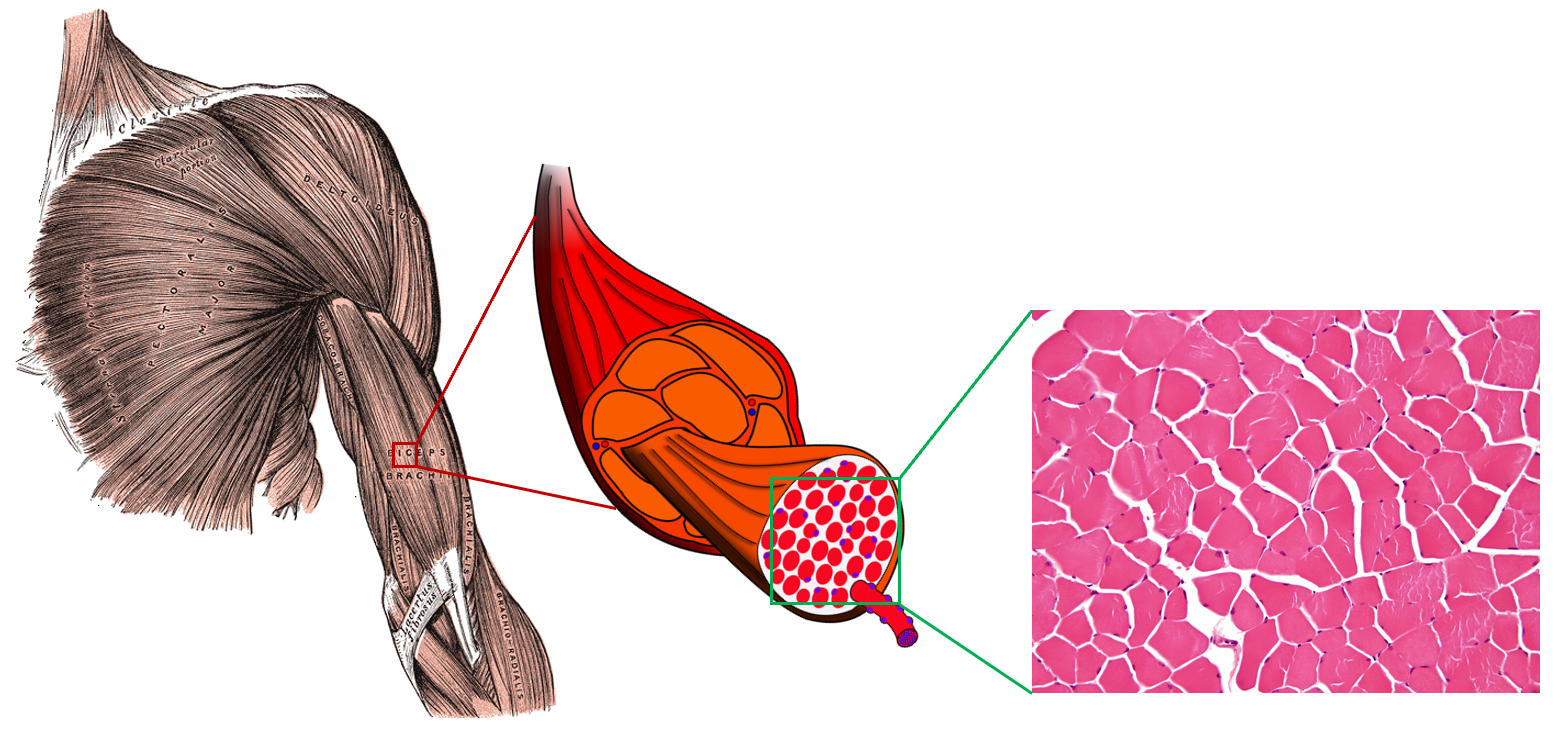
My PhD research focus was on non-invasively measuring the microstructure (or cellular organization) of skeletal muscle. Muscle microstructure consists of long fibers that are tightly packed together and surrounded by an extracellular collagen matrix. Changes in this microstructure are related to changes in muscle health, so non-invasive measurement of this microstructure should provide new diagnostic tools for assessing muscle health.
I work on measuring this microstructure using diffusion-weighted magnetic resonance imaging (dMRI) and connecting this microstructure to effective mechanical properties of muscle. This work can be broadly organized as two interrelated problems: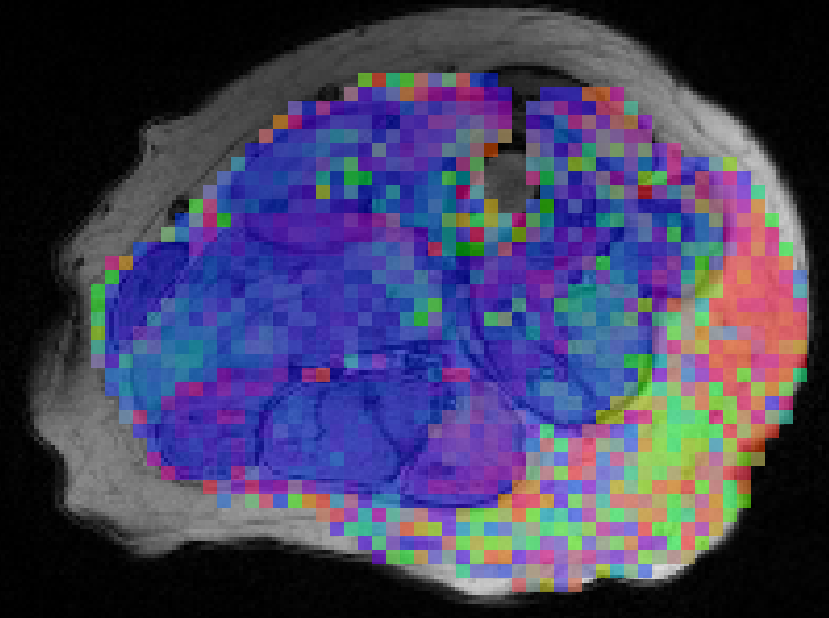
The Forward Problem: Understanding how changes in skeletal muscle microstructure affect the dMRI signal.
The Inverse Problem: Estimating the microstructure of skeletal muscle from dMRI measurements.
By solving these problems, I am working to create an imaging pipeline that can image a skeletal muscle, estimate its microstructure, and use this microstructural information to estimate key properties of the muscle. This pipeline will better inform our understanding of muscle’s functional ability. Much of this work is based on measuring the muscle’s microstructure using dMRI. dMRI measures the movement of water molecules. By measuring how microstructural restrictions affect the movement of these water molecules, we can measure the cumulative effect of the microstructure on the dMRI signal. For a brief introduction into how we can measure muscle microstructure using dMRI, check out the video below.
Forward Problem
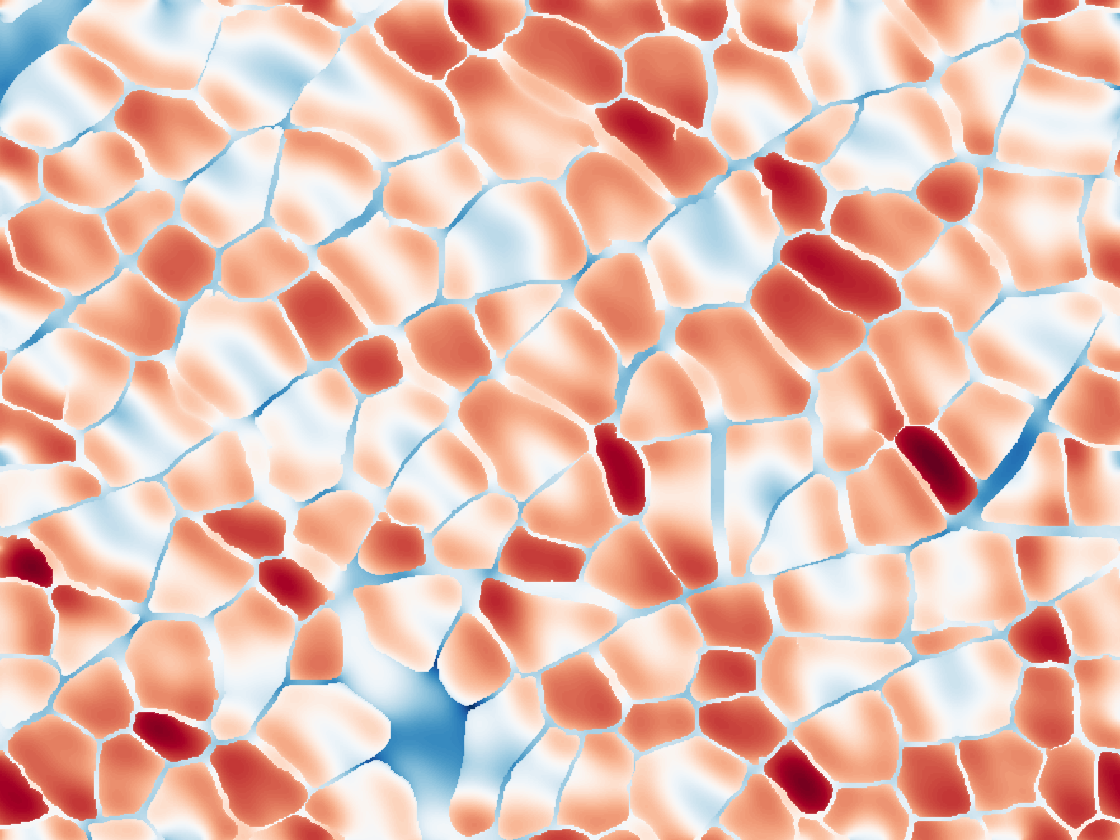
How do changes in skeletal muscle microstructure affect the dMRI signal? In order to predict microstructure with dMRI, we first need to answer this question. To answer it, we perform numerical simulations of dMRI in skeletal muscle using both realistic and simplified geometric models. These simulations allow us to understand how the dMRI signal will change when there is a change in the underlying microstructure. We call this the forward problem because it involves solving a problem that mimics what actually happens in a dMRI experiment, where the underlying structure affects the measured signal.
learn more
Inverse Problem
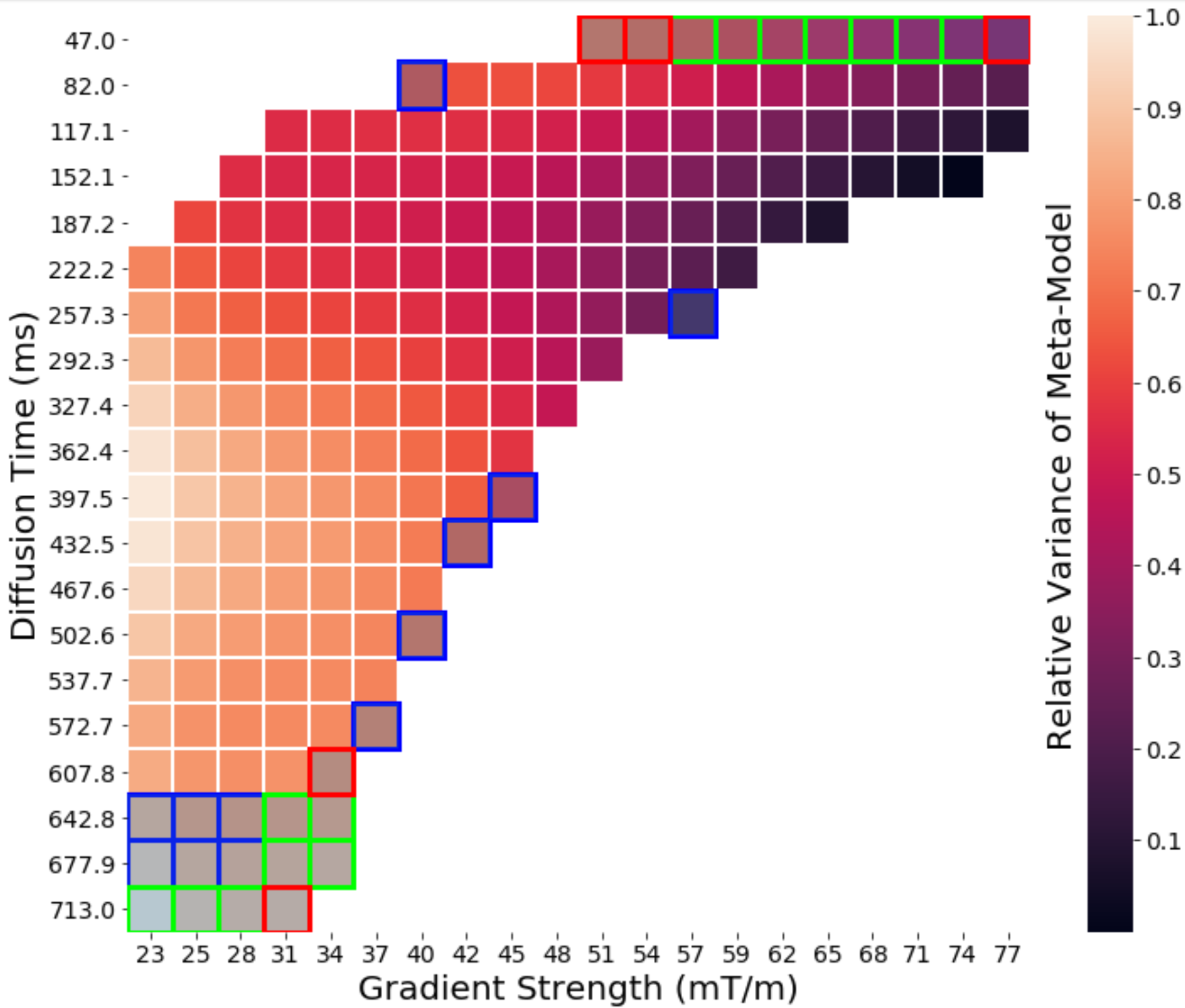
The inverse problem asks “how do you take the dMRI signal and determine the microstructure of the muscle from it?” We use the insights we gain from solving the forward problem to invert the forward problem by taking the measured dMRI signal and going backwards to the original microstructure that caused the signal. These models consist of analytical compartment models, numerical inversion methods, meta-models and machine learning techniques. They allow us to determine what dMRI pulse sequences encode the most information into the signal and how to decode this information to recover the underlying microstructure.